Note: The term Haplogroup and Haplotype are strictly defined in the biological sciences. But a bit misappropriated by the genetic genealogy community. Any definition here is subject to criticism; depending on your background and view. Needless to say, we try to define a more strict definition that seems to be the more common use in the genetic genealogy community.
A Haplogroup in genetics is defined as a group of individuals with a common set of shared alleles. Specifically, shared derived SNPs that different than in the referenced human genome model. A member of a haplogroup is "positive for change" in all the SNPs that define the haplogroup (that is, they have all the alleles). Occasionally, if not all SNPs are reliably tested, they may be presumed derived based on other information. If an SNP in a haplogroup is measured as ancestral when all the rest are derived, then either the tester is forming a new branch that splits the haplogroup (more rare) or the test result is likely considered incorrect (more common). In even rarer cases, the haplogroup and tree it resides in could be mis-formed. Defining haplogroups and ordering them in time is what developing a phylogenetic tree is all about.
There may be only a single SNP that defines a haplogroup; but often there are more. Sometimes tens to maybe a hundred or more. An SNP is defined by the chromosome, position (coordinate) and value change from the reference model. An SNP may have one or more names.
Different SNPs that occur in the same haplogroup are phylogenetically "equivalent". Some in the genetic genealogy community shorten this to simply equivalent. Each phylogenetically "equivalent" SNP is unique in name an definition in the haplogroup.
Sometimes the same SNP has been named by different organizations using different names but that are identical in all other aspects. These are "aliases" for the same SNP. They are not equivalent SNPs. But identical in every respect except the name. Identical SNPs that have different names are aliases for each other. The SNP is considered defined only once for the haplogroup even though the aliases may all be included.
Sometimes you may see names like L176, L176.1, L176.2 and so on. These are the same SNP in all aspects except the name. Often only differing in name with a suffix of a dot (.) and number. But unlike aliases in the same haplogroup, these recurrent SNPs are placed in different haplogroups. Like the name implies, this SNP has happened more than once in the history of mankind. But at different times and different branches of the ancestral phylogenetic tree. To avoid confusion, they are given unique names so every SNP in every haplogroup is uniquely named in the overall phylogenetic tree. These can be distinguished from aliases in that the similar names are in different haplogroups. Aliases always occur in the same haplogroup.
Note that most SNPs, even when outside yDNA and mtDNA, have an rsID from dbSNP assigned to them which defines them fully. Identical in the above means SNPs with the same DNA strand, same coordinate and name, Unique means not Identical.
Different tree creators may use a different SNPs in a haplogroup to name what is essentially the same haplogroup.
When places in a phylogenetic tree, a haplogroup can have an estimated century of entry and one of exit. Essentially giving a time range for the haplogroup. The times are really associated with the branches leading into and out of the haplogroup. But often reported with the haplogroup in the phylogenetic tree.
There is no known order of creation for phylogenetically equivalent SNPs. Each equivalent SNP is unique and different. Only when a haplogroup is reduced to a single SNP, can the date range be attached to the SNP itself by virtue of its placement in the phylogenetic tree.
When named by one of the "equivalent" SNPs, this is termed the YCC short notation naming.
Some haplogroups at the top level of a phylogenetic tree, are named using the original long form ((YCC) naming convention. Sometimes, a top level. major branch in the phylogenetic tree may be used to additionally qualify a haplogroup name. So as to make it a little more recognizable and defined. Even possibly giving a partial, pseudo long-form YCC name of major branch points to get to it. Sometimes the haplogroup definition will give the aliases for the same SNP. Others only one; often the name they may have created. Aliases occur when publication of research work is delayed and somewhat in parallel. Possibly also when the creator of the name has not done their homework to discover if it was already named.
Note that a recurrent SNP does not represent a different change in value (allele) nor a change back to the ancestral value. They would be named uniquely and differently even though sharing the same coordinate in the DNA strand.
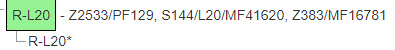
To understand this all a bit better, lets look at haplogroup R-L20 as shown here. In the example, a haplogroup named R-L20 has 3 defined SNPs that form it. They are each separated by commas here. Each of these SNPs has one or more aliases for it. So SNP Z2533 also has an alias PF129. And S144 has the more commonly known alias of L20. Which is the name and which is the alias for an SNP is arbitrary and use sensitive.
Some phylogenetic tree designers use the lowest numbered SNP (or alias) to name the haplogroup. Some the lowest letter designator. Others their own, if named by them. Each tree designer has their own choice. Just as the creation and management of their phylogenetic tree is their choice. There are many tree creators and thus many forms of haplogroup naming and designation. A haplogroup can often be called a "block" in a phylogenetic tree. In a strict sense, they are edges in the graph. But in most modern trees, they are the nodes or branch points. And hence represented as blocks at those nodes with the information defining them.
Historically, haplogroups were defined for ancient, anthropological-defined human populations. They only came into genetic genealogy use to help verify that those with similar haplotypes were also in the same haplogroup and thus truly matching and part of the same patriline. Often, testers have, a predicted haplogroup from a haplotype. But then expansion in personal genomic testing of SNPs allowed for direct testing of haplogroups. More recently, due to full sequence testing, much more extensive discoveries of novel SNPs are being made. And as a result, haplogroups are being defined in the genealogical time frame for particular surname lines (i.e. family branches). Thus pushing haplogroups and their corresponding sub-clades into the mainstream genetic genealogy process. And sometimes showing how haplotypes can be in error due to convergence or other issues with the more rapid change in STR values.
Often an individual belongs to many haplogroups. This because a haplogroup is a proper subset of all the known derived SNPs. An allele only appears in a single haplogroup unless recurrent. But a named allele will be unique among all haplogroups.
Deeper studies try to predict or determine when a haplogroup formed in the past. Or at least an order to the SNP changes between haplogroups can be made on time. This ordering (more properly, a mathematical partial order) of haplogroup definitions is then termed a phylogenetic tree (and is more properly, a mathematical spanning tree with a defined root) When you hear the term haplogroup tree or even haplotree, they are really referring more properly to a phylogenetic tree of haplogroups. We do not use these shortened terms here.
Individuals are usually identified with a single leaf haplogroup, paragroup, or sometimes intermediate haplogroup. Usually the lowest / deepest / most-recent-in-time / farthest-from-the-root in the phylogenetic tree they have had SNPs tested for that show as derived. If fully tested, often a leaf or paragroup in the tree. Sometimes designated with an asterisk to mean tested for the clade but negative for all the subclades. Possibly ancient in the tree itself. You should be reminded that a vast majority of the tree is developed from testing recent, living individuals. So a rare SNP value may be a novel below a current leaf, or possibly part of the root or very ancient haplogroup that they branch from and virtually no one else does, or something in between. Just because a value is rare does not mean it is very recent in occurrence.
Depending on the level of testing performed, a testers haplogroup may not "match" a near relative. In such cases, you need to view the phylogenetic tree and see if one haplogroup is in a subclade of the other. That is, deeper / lower / more-recent-in-time in the phylogenetic tree than the other; or one haplogroup is on the path down to it from the root of the tree through the other haplogroup. If so, then the testers have a common shared ancestral haplogroup and could possibly share a deeper haplogroup on the tree if they both tested to the same level.
Often, microarray tests will only test SNPs that exist in ancient haplogroups. Often only one in that haplogroup. So a user is designated as being in the haplogroup of the deepest haplogroup showing a derived SNP value for with other SNPs in that haplogroup being presumed derived also (as well as any intermediary SNPs in earlier haplogroups not tested).
Generally, haplogroups in humans are only defined for particular alleles on the yDNA and mtDNA strands. This because that DNA does not recombine and is stable for thousands of generations (tens of thousands of years). But who knows. As the field of genealogy and genetic anthropology intertwine even more, maybe they can start identifying autosomal and xDNA alleles belonging to a stable haplogroup also. After all, that is what the ad-mixture analysis (aka ethnicity charts) is all about. Identifying haplogroups of autosomal alleles that are unique to a population or area.
A Haplogroup in genetics is defined as a group of individuals with a common set of shared alleles. Specifically, shared derived SNPs that different than in the referenced human genome model. A member of a haplogroup is "positive for change" in all the SNPs that define the haplogroup (that is, they have all the alleles). Occasionally, if not all SNPs are reliably tested, they may be presumed derived based on other information. If an SNP in a haplogroup is measured as ancestral when all the rest are derived, then either the tester is forming a new branch that splits the haplogroup (more rare) or the test result is likely considered incorrect (more common). In even rarer cases, the haplogroup and tree it resides in could be mis-formed. Defining haplogroups and ordering them in time is what developing a phylogenetic tree is all about.
Equivalent, Alias and Recurrent SNPs
There are some terms that categorize or refine an SNP that are strictly related to haplogroups and phylogenetic trees.There may be only a single SNP that defines a haplogroup; but often there are more. Sometimes tens to maybe a hundred or more. An SNP is defined by the chromosome, position (coordinate) and value change from the reference model. An SNP may have one or more names.
Different SNPs that occur in the same haplogroup are phylogenetically "equivalent". Some in the genetic genealogy community shorten this to simply equivalent. Each phylogenetically "equivalent" SNP is unique in name an definition in the haplogroup.
Sometimes the same SNP has been named by different organizations using different names but that are identical in all other aspects. These are "aliases" for the same SNP. They are not equivalent SNPs. But identical in every respect except the name. Identical SNPs that have different names are aliases for each other. The SNP is considered defined only once for the haplogroup even though the aliases may all be included.
Sometimes you may see names like L176, L176.1, L176.2 and so on. These are the same SNP in all aspects except the name. Often only differing in name with a suffix of a dot (.) and number. But unlike aliases in the same haplogroup, these recurrent SNPs are placed in different haplogroups. Like the name implies, this SNP has happened more than once in the history of mankind. But at different times and different branches of the ancestral phylogenetic tree. To avoid confusion, they are given unique names so every SNP in every haplogroup is uniquely named in the overall phylogenetic tree. These can be distinguished from aliases in that the similar names are in different haplogroups. Aliases always occur in the same haplogroup.
| Name | |
| Equivalent | Unique SNPs in the same haplogroup |
| Alias | Identical SNPs with different names in the same haplogroup |
| Recurrent | Identical SNPs with slightly different names in different haplogroups |
| Un-named | SNPs not (yet) assigned a name; identified by 1-43629:C>T or similar |
Note that most SNPs, even when outside yDNA and mtDNA, have an rsID from dbSNP assigned to them which defines them fully. Identical in the above means SNPs with the same DNA strand, same coordinate and name, Unique means not Identical.
Naming Haplogroups
One of the phylogenetically equivalent SNP is most often used to name the haplogroup today. SNPs and the haplogroups they reside in are often named when added to a phylogenetic tree.Different tree creators may use a different SNPs in a haplogroup to name what is essentially the same haplogroup.
When places in a phylogenetic tree, a haplogroup can have an estimated century of entry and one of exit. Essentially giving a time range for the haplogroup. The times are really associated with the branches leading into and out of the haplogroup. But often reported with the haplogroup in the phylogenetic tree.
There is no known order of creation for phylogenetically equivalent SNPs. Each equivalent SNP is unique and different. Only when a haplogroup is reduced to a single SNP, can the date range be attached to the SNP itself by virtue of its placement in the phylogenetic tree.
When named by one of the "equivalent" SNPs, this is termed the YCC short notation naming.
Some haplogroups at the top level of a phylogenetic tree, are named using the original long form ((YCC) naming convention. Sometimes, a top level. major branch in the phylogenetic tree may be used to additionally qualify a haplogroup name. So as to make it a little more recognizable and defined. Even possibly giving a partial, pseudo long-form YCC name of major branch points to get to it. Sometimes the haplogroup definition will give the aliases for the same SNP. Others only one; often the name they may have created. Aliases occur when publication of research work is delayed and somewhat in parallel. Possibly also when the creator of the name has not done their homework to discover if it was already named.
Note that a recurrent SNP does not represent a different change in value (allele) nor a change back to the ancestral value. They would be named uniquely and differently even though sharing the same coordinate in the DNA strand.
Haplogroup Example
Haplogroup Example
Some phylogenetic tree designers use the lowest numbered SNP (or alias) to name the haplogroup. Some the lowest letter designator. Others their own, if named by them. Each tree designer has their own choice. Just as the creation and management of their phylogenetic tree is their choice. There are many tree creators and thus many forms of haplogroup naming and designation. A haplogroup can often be called a "block" in a phylogenetic tree. In a strict sense, they are edges in the graph. But in most modern trees, they are the nodes or branch points. And hence represented as blocks at those nodes with the information defining them.
Paragroups
A haplogroup that is named with a trailing asterisk ('*') is special and termed a paragroup. It is a subclade to capture all the testers that are not in any other subclade (so negative or ancestral for all lower SNPs in subclades). They likely still has some unique SNPs not captured in the tree yet. So a paragroup member will likely be in a new (not yet defined) subclade. Just awaiting for more testers in their line to be able to define the branching. Generally, a tree awaits two solid confirmed cases of testers with similar derived, novel SNP values before forming a new sub-branch based on those SNPs.Historically, haplogroups were defined for ancient, anthropological-defined human populations. They only came into genetic genealogy use to help verify that those with similar haplotypes were also in the same haplogroup and thus truly matching and part of the same patriline. Often, testers have, a predicted haplogroup from a haplotype. But then expansion in personal genomic testing of SNPs allowed for direct testing of haplogroups. More recently, due to full sequence testing, much more extensive discoveries of novel SNPs are being made. And as a result, haplogroups are being defined in the genealogical time frame for particular surname lines (i.e. family branches). Thus pushing haplogroups and their corresponding sub-clades into the mainstream genetic genealogy process. And sometimes showing how haplotypes can be in error due to convergence or other issues with the more rapid change in STR values.
Often an individual belongs to many haplogroups. This because a haplogroup is a proper subset of all the known derived SNPs. An allele only appears in a single haplogroup unless recurrent. But a named allele will be unique among all haplogroups.
Deeper studies try to predict or determine when a haplogroup formed in the past. Or at least an order to the SNP changes between haplogroups can be made on time. This ordering (more properly, a mathematical partial order) of haplogroup definitions is then termed a phylogenetic tree (and is more properly, a mathematical spanning tree with a defined root) When you hear the term haplogroup tree or even haplotree, they are really referring more properly to a phylogenetic tree of haplogroups. We do not use these shortened terms here.
Haplogroups versus Clades and Subclades
A subgroup, or subclade as it is usually termed, is simply a specific branch down-from a specified haplogroup in the phylogenetic tree. That is, pick a haplogroup in the tree to be a new root. Then each haplogroup immediately below it is a subclade. The base and all its subclade(s) are termed a clade. So a clade is defined by the root haplogroup and forms its own phylogenetic tree.Individuals are usually identified with a single leaf haplogroup, paragroup, or sometimes intermediate haplogroup. Usually the lowest / deepest / most-recent-in-time / farthest-from-the-root in the phylogenetic tree they have had SNPs tested for that show as derived. If fully tested, often a leaf or paragroup in the tree. Sometimes designated with an asterisk to mean tested for the clade but negative for all the subclades. Possibly ancient in the tree itself. You should be reminded that a vast majority of the tree is developed from testing recent, living individuals. So a rare SNP value may be a novel below a current leaf, or possibly part of the root or very ancient haplogroup that they branch from and virtually no one else does, or something in between. Just because a value is rare does not mean it is very recent in occurrence.
Depending on the level of testing performed, a testers haplogroup may not "match" a near relative. In such cases, you need to view the phylogenetic tree and see if one haplogroup is in a subclade of the other. That is, deeper / lower / more-recent-in-time in the phylogenetic tree than the other; or one haplogroup is on the path down to it from the root of the tree through the other haplogroup. If so, then the testers have a common shared ancestral haplogroup and could possibly share a deeper haplogroup on the tree if they both tested to the same level.
Often, microarray tests will only test SNPs that exist in ancient haplogroups. Often only one in that haplogroup. So a user is designated as being in the haplogroup of the deepest haplogroup showing a derived SNP value for with other SNPs in that haplogroup being presumed derived also (as well as any intermediary SNPs in earlier haplogroups not tested).
Generally, haplogroups in humans are only defined for particular alleles on the yDNA and mtDNA strands. This because that DNA does not recombine and is stable for thousands of generations (tens of thousands of years). But who knows. As the field of genealogy and genetic anthropology intertwine even more, maybe they can start identifying autosomal and xDNA alleles belonging to a stable haplogroup also. After all, that is what the ad-mixture analysis (aka ethnicity charts) is all about. Identifying haplogroups of autosomal alleles that are unique to a population or area.
Major yDNA Haplogroup and Phylogenetic Tree Sites
- ISOGG: ISOGG Tree
- yFull: yFull Tree
- yTree: yTree Tree (only Haplogroup R1b-P312)) and below)
- FamilyTreeDNA: (Big Y tree only available by login to those BigY tested on that site; less detailed public tree otherwise)
- NGG: (only available by login to those tested on that site) (see Trees (archived) for a hint) (went away in 2020)
Major mtDNA Haplogroup and Phylogenetic Tree Sites
- PhyloTree: Tree
- FamilyTreeDNA: (only available by login to those tested on that site; less detailed public tree otherwise)
- yFull: yFull mTree (in active development due to WGS test submissions)
- NGG: (only available by login to those tested on that site) (Trees (archived) for a hint) (went away in 2020)
External References
- The YCC Consortium, A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups, Genome Res. 2002. 12: 339-348
- 23andMe tutorial
- SNPedia on Y and mt haplogroups
- Eupedia article on general background of haplogroups
- Wikipedia definition of Haplogroup and Paragroup
- ISOGG definition